Data
We believe in openly sharing the major datasets supporting our work. Most of our publications and preprints should already contain a section detailing where and how the relevant data is available. For Code or Reagents, which we will also freely share, please see the other relevant sections.
Most of the imaging data will be available in the form of trajectories and the genomics data is available in the standard formats. If you have any trouble locating a dataset or believe one to be missing, please Contact Us, and we will make sure to get back to you.
Most of the imaging data will be available in the form of trajectories and the genomics data is available in the standard formats. If you have any trouble locating a dataset or believe one to be missing, please Contact Us, and we will make sure to get back to you.
Click on the relevant publication name for more information:
Region Capture Micro-C reveals coalescence of enhancers and promoters into nested microcompartments
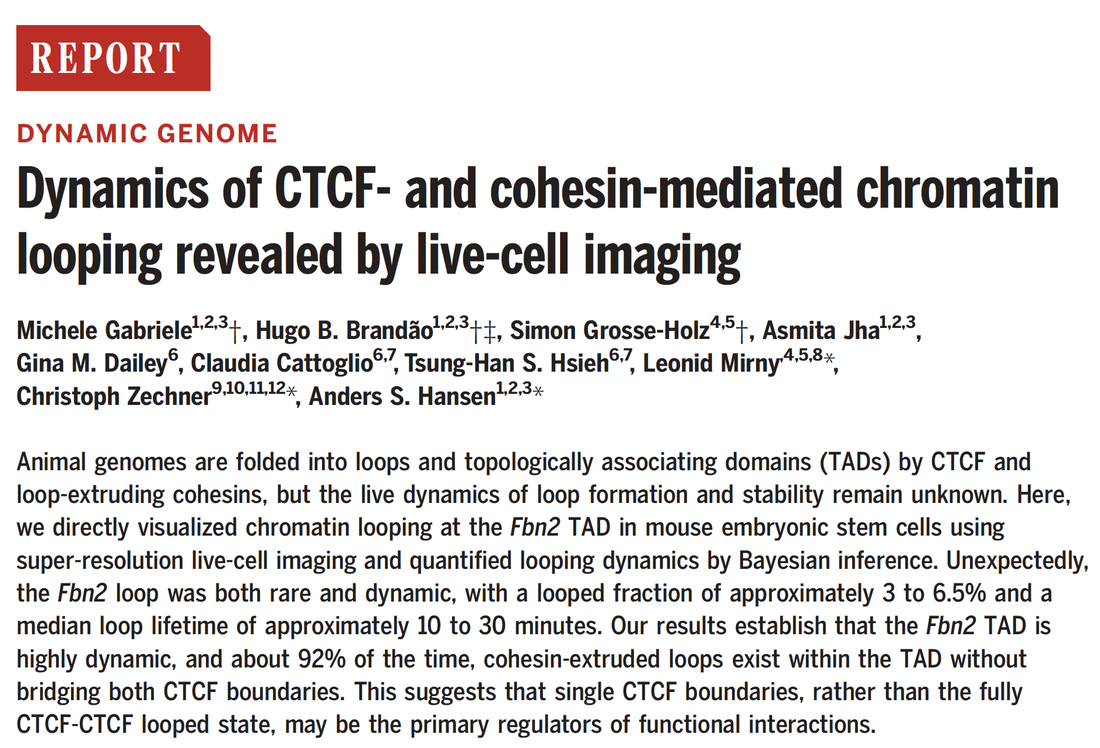
Dynamics of CTCF- and cohesin-mediated chromatin looping revealed by live-cell imaging
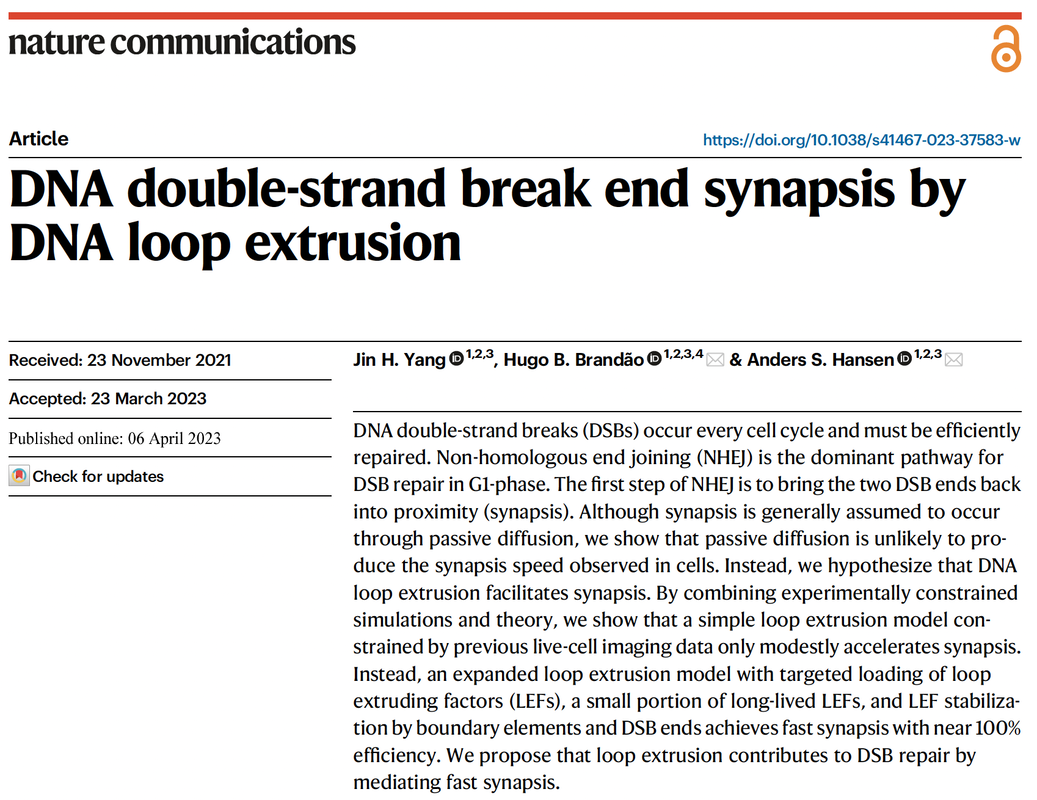
DNA double-strand break end synapsis by DNA loop extrusion
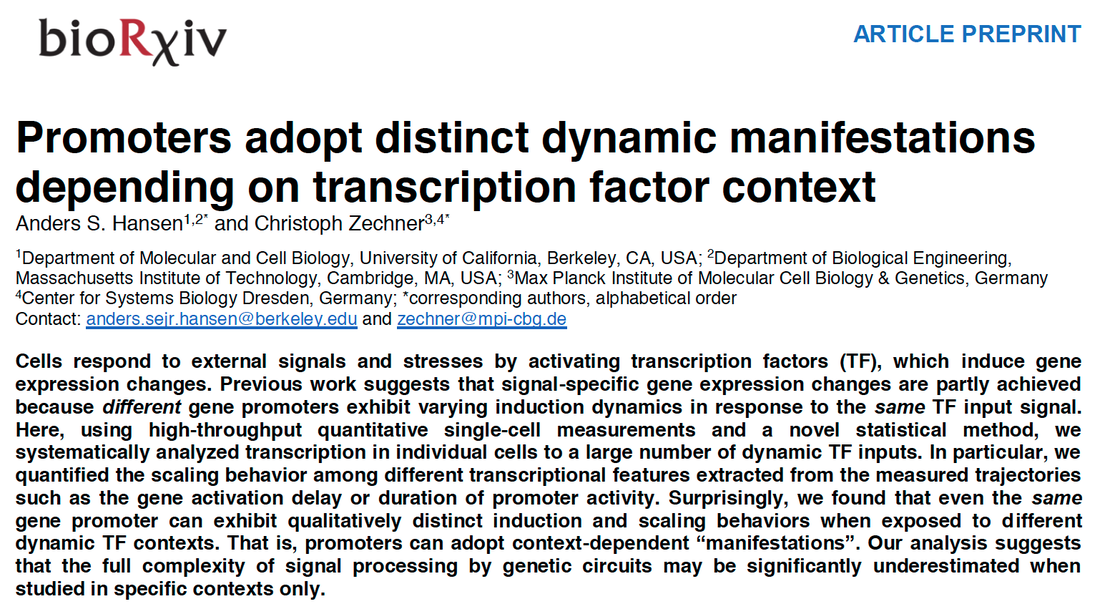
Promoters adopt distinct dynamic manifestations depending on transcription factor context
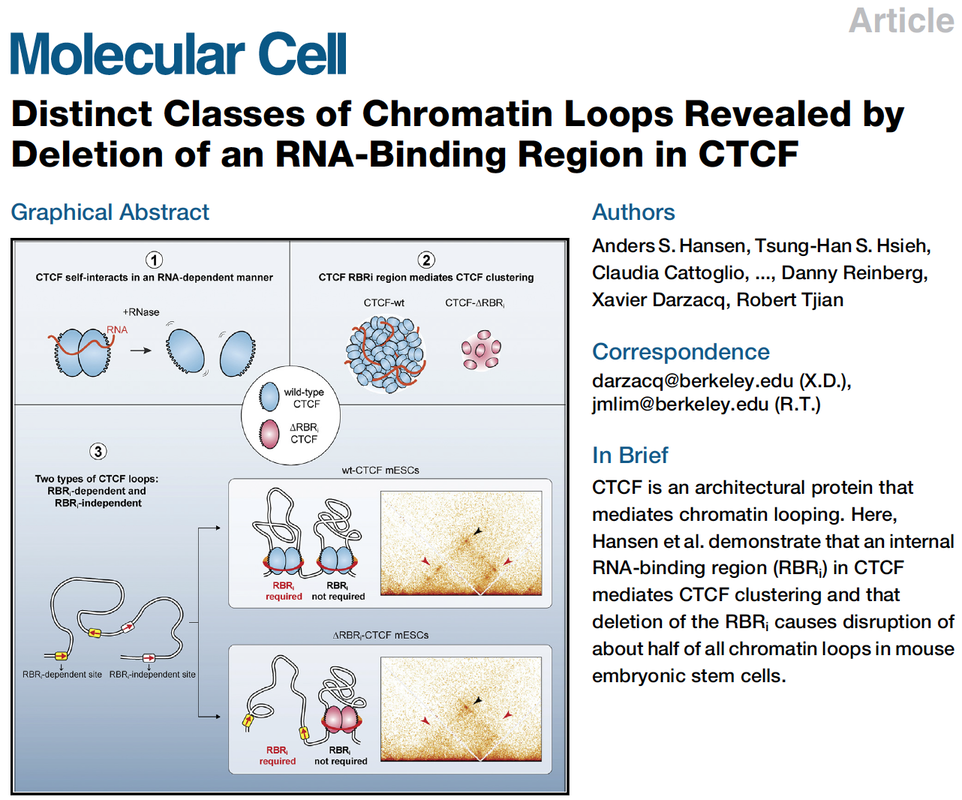
An RNA-binding region in CTCF regulates clustering and chromatin looping
Guided nuclear exploration increases CTCF target search efficiency
Determining cellular CTCF and cohesin abundances to constrain 3D genome models
Evidence for DNA-mediated nuclear compartmentalization distinct from phase separation
CTCF sites display cell cycle dependent dynamics in factor binding and nucleosome positioning
RNA polymerase II clustering through carboxy-terminal domain phase separation
Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II
Robust model-based analysis of single-particle tracking experiments with Spot-On
CTCF and Cohesin Regulate Chromatin Loop Stability with Distinct Dynamics
cis Determinants of Promoter Threshold and Activation Timescale
Limits on information transduction through amplitude and frequency regulation of transcription factor activity
Promoter decoding of transcription factor dynamics involves a trade-off between noise and control of gene expression
Region Capture Micro-C reveals coalescence of enhancers and promoters into nested microcompartments
Dynamics of CTCF- and cohesin-mediated chromatin looping revealed by live-cell imaging
DNA double-strand break end synapsis by DNA loop extrusion
Promoters adopt distinct dynamic manifestations depending on transcription factor context
An RNA-binding region in CTCF regulates clustering and chromatin looping
Guided nuclear exploration increases CTCF target search efficiency
Determining cellular CTCF and cohesin abundances to constrain 3D genome models
Evidence for DNA-mediated nuclear compartmentalization distinct from phase separation
CTCF sites display cell cycle dependent dynamics in factor binding and nucleosome positioning
RNA polymerase II clustering through carboxy-terminal domain phase separation
Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II
Robust model-based analysis of single-particle tracking experiments with Spot-On
CTCF and Cohesin Regulate Chromatin Loop Stability with Distinct Dynamics
cis Determinants of Promoter Threshold and Activation Timescale
Limits on information transduction through amplitude and frequency regulation of transcription factor activity
Promoter decoding of transcription factor dynamics involves a trade-off between noise and control of gene expression
Region Capture Micro-C reveals coalescence of enhancers and promoters into nested microcompartments
Goel VY, Huseyin MK, Hansen AS^. Region Capture Micro-C reveals coalescence of enhancers and promoters into nested microcompartments. Nature Genetics, 2023. The raw data and code is freely available:
|
Dynamics of CTCF- and cohesin-mediated chromatin looping revealed by live-cell imaging
Gabriele M*, Brandão HB*, Grosse-Holz S*, Jha A, Dailey GM, Cattoglio C, Hsieh THS, Mirny L^, Zechner C^, Hansen AS^. Dynamics of CTCF and cohesin mediated chromatin looping revealed by live-cell imaging. Science, 2022. |
DNA double-strand break end synapsis by DNA loop extrusion
Yang, HY, Brandão HB, Hansen AS. DNA double-strand break end synapsis by DNA loop extrusion. Nature Communications, 2023 Please see the links below for the raw data:
|
Promoters adopt distinct dynamic manifestations depending on transcription factor context
Hansen AS^; Zechner C^. Promoters adopt distinct dynamic manifestations depending on transcription factor context. bioRxiv, 2019, 650762. Please see the links below for the raw data:
|
Distinct classes of chromatin loops revealed by deletion of an RNA-binding region in CTCF
Hansen AS*; Hsieh THS*; Cattoglio C*; Pustova I; Saldana-Meyer R; Reinberg D; Darzacq X^; Tjian R^. Distinct Classes of Chromatin Loops Revealed by Deletion of an RNA-Binding Region in CTCF. Molecular Cell, 2019, DOI:https://doi.org/10.1016/j.molcel.2019.07.039. |
Guided nuclear exploration increases CTCF target search efficiency
Hansen AS*; Amitai A*; Cattoglio C; Tjian R; Darzacq X. Guided nuclear exploration increases CTCF target search efficiency. bioRxiv, 2018, 495457. For this paper, we conducted 1693 spaSPT experiments (so single-molecule tracking experiments in 1693 single cells). All the raw trajectory data is freely available on Zenodo and all the code is available from GitLab:
|
Determining cellular CTCF and cohesin abundances to constrain 3D genome models
Cattoglio C; Pustova I; Walther N; Ho JJ; Hantsche-Grininger M; Inouye CJ; Hossain MJ; Dailey GM; Ellenberg J; Darzacq X; Tjian R; Hansen AS: Determining cellular CTCF and cohesin abundances to constrain 3D genome models. eLife 2019, 40164.
|
Transient DNA binding induces RNA Polymerase II compartmentalization during Herpesviral infection distinct from phase separation
McSwiggen DT; Hansen AS; Marie-Nelly H; Teves SS; Heckert A; Hao Y; Umemoto K; Dugast-Darzacq C; Tjian R^; Darzacq X^. Evidence for DNA-mediated nuclear compartmentalization distinct from phase separation. eLife 2019, e47098. |
CTCF sites display cell cycle dependent dynamics in factor binding and nucleosome positioning
Oomen ME; Hansen AS; Liu Y; Darzacq X; Dekker J. CTCF sites display cell cycle dependent dynamics in factor binding and nucleosome positioning. Genome Research 2019 29, 236-249 All the raw SPT data for this paper is freely available on Zenodo. The data was analyzed with Spot-On Matlab (please see the paper for full details on parameters), which is freely available from GitLab.
|
RNA polymerase II clustering through carboxy-terminal domain phase separation
Boehning M; Dugast-Darzacq C; Rankovic M; Hansen AS; Yu TK; Marie-Nelly H; Kokic G; Dailey GM; Cramer P; Darzacq X; Zweckstetter M. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nature Structural and Molecular Biology 2018, 25, 833–840. The raw PALM and spaSPT data is available at Zenodo. The spaSPT data was analyzed with Spot-On Matlab (please see the paper for full details on parameters), which is freely available from GitLab.
|
Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II
Lu H; Yu D; Hansen AS; Ganguly S; Liu R; Heckert A; Darzacq X; Zhou Q; 2018. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 2018 558, 318–323 All the raw SPT data for this paper is freely available on Zenodo. The data was analyzed with Spot-On Matlab (please see the paper for full details on parameters), which is freely available from GitLab.
|
Robust model-based analysis of single-particle tracking experiments with Spot-On
Hansen AS*; Woringer M*; Grimm J; Lavis LD; Tjian R; Darzacq X. Robust model-based analysis of single-particle tracking experiments with Spot-On. eLife 2018 e33125 For this paper, we conducted 1064 spaSPT experiments (so single-molecule tracking experiments in 1064 single cells). All the raw trajectory data is freely available on Zenodo and all the code is available from GitLab:
|
CTCF and Cohesin Regulate Chromatin Loop Stability with Distinct Dynamics
Hansen AS; Pustova I; Cattoglio C; Tjian R; Darzacq X. CTCF and Cohesin Regulate Chromatin Loop Stability with Distinct Dynamics. eLife 2017. e25776
|
Cis-determinants of promoter threshold and activation timescale
Hansen AS; O’Shea EK. Cis-determinants of promoter threshold and activation timescale. Cell Reports 2015, 12(8), 1226-1233
|
Limits on information transduction through amplitude and frequency regulation of transcription factor activity
Hansen AS; O’Shea EK. Limits on information transduction through amplitude and frequency regulation of transcription factor activity. eLife 2015 e06559
|
Promoter decoding of transcription factor dynamics involves a trade-off between noise and control of gene expression
Hansen AS; O’Shea EK. Promoter decoding of transcription factor dynamics involves a trade-off between noise and control of gene expression. Molecular Systems Biology 2013, 9.
|