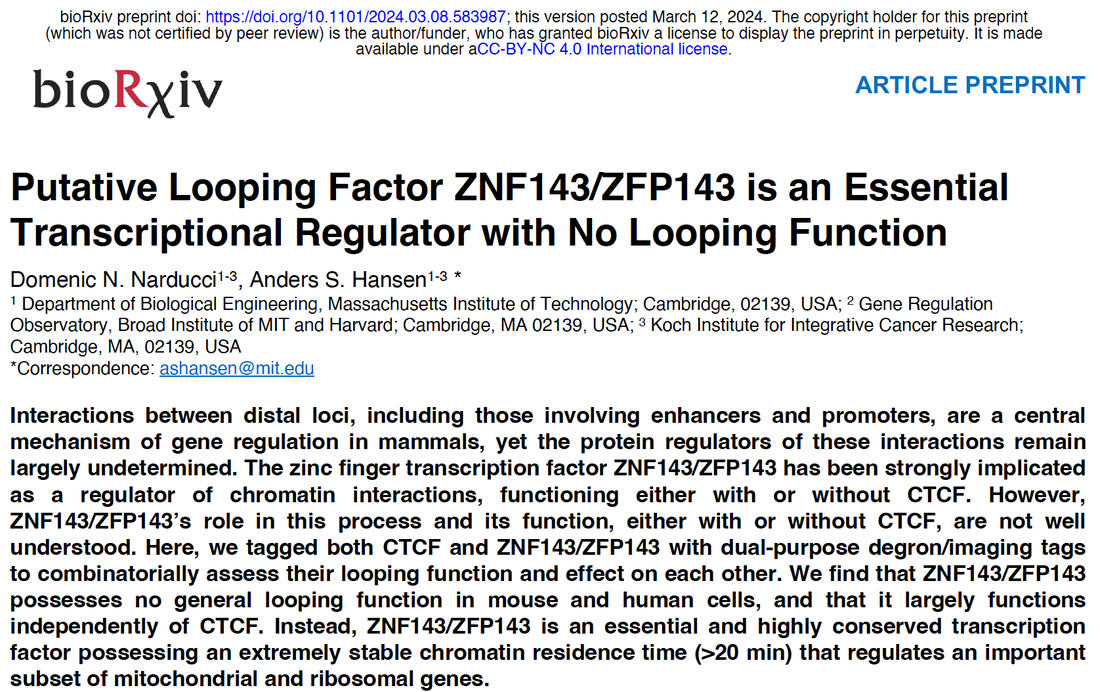
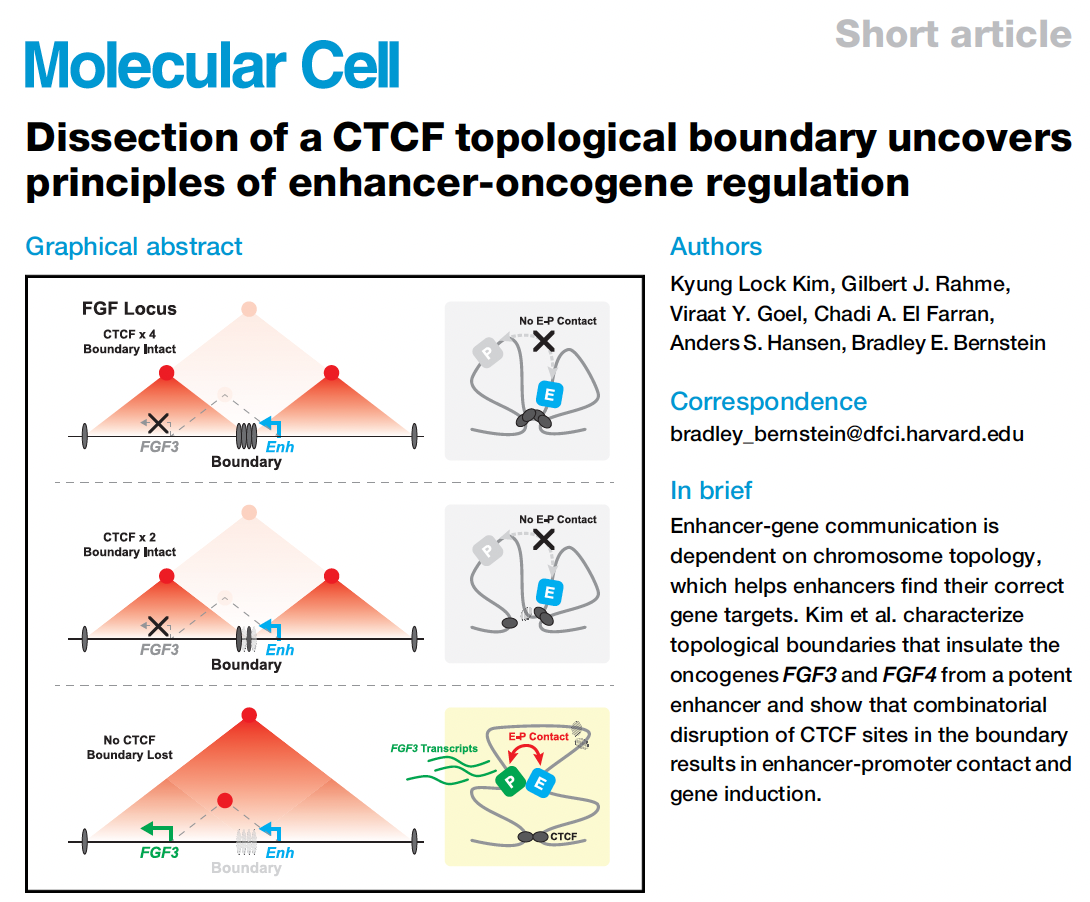
Congrats to Viraat for RCMC contributions to a new paper led by KL Kim and Brad Bernstein, where we used RCMC to understand how partial and full loss of a CTCF insulator regulates enhancer-oncogene interaction strength and oncogene expression in GIST cancer. You can find the full paper in Molecular Cell.
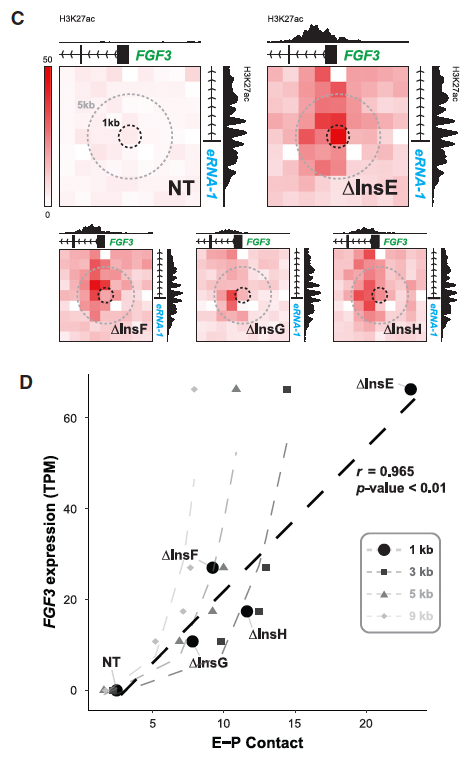
A neat part of the paper is Fig. 4C,D which really highlights that the enhancer-promoter vs. expression analysis can be either linear or non-linear (convex) depending on the resolution at which the chromosome conformation capture data is analyzed. Congratulations to Harvey on the publication of his new review article in Nature Reviews Molecular Cell Biology, which can be found here.
We went to a Celtics game and celebrated! Good news this week:
We got a new "Bridge Grant" with Zuzana Tothova (Dana-Farber) to study the effect of loss of function mutations in cohesin that cause cancer. Also Anders has a new title: Associate Professor. The lab turned 4 years old today! We celebrated 4 fantastic years at SPIN Boston with a few rounds of table tennis.
We are very happy to welcome Dominque Dang and Sophie Wang, both first year undergraduates, who have joined the lab as UROPs during IAP. Dom is working with Sarah on bidirectional promoters and Sophie is working with Dominic on integrated FRAP and SPT analysis.
Thanks to the NSF for awarding us with a CAREER award that will run until 2029 to support our research into loop extrusion and transcription, as well as the design of a new teaching module and outreach activities.
We are thrilled to welcome 3 new lab members:
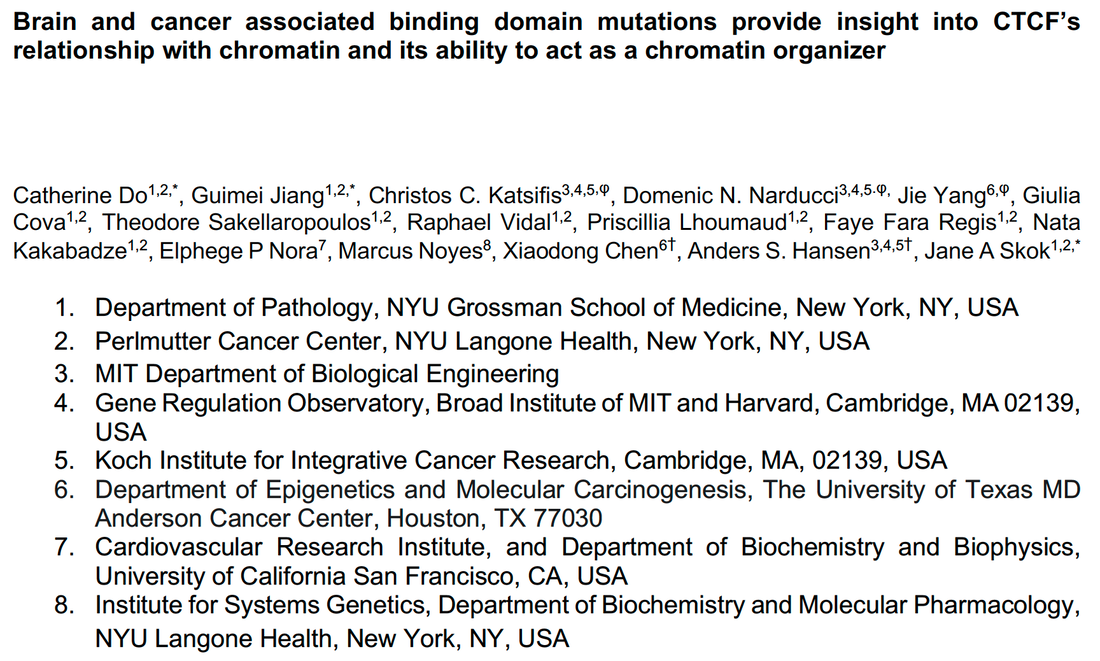
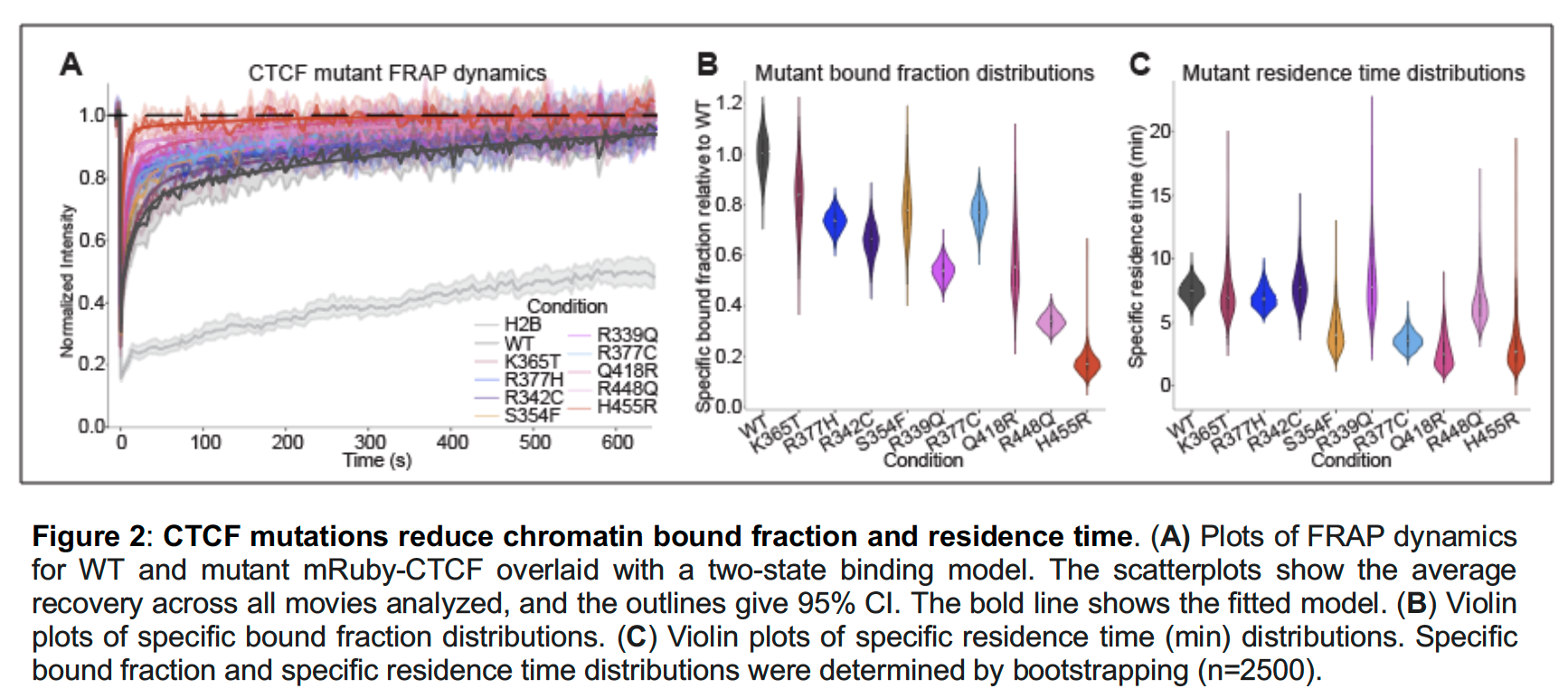
Allison Vieira has just joined the lab as a tech associate and will be managing the lab and working to understand genome structure and function. Michael Quezada is a Harvard-MIT MD-PhD graduate student working jointly with Jason Buenrostro and working on understanding the selectivity of intrinsically disordered regions at scale Varshini Ramanathan is a BE graduate student and will work on loops, bioinformatically finding them and watching them form as cells respond to their environment. Welcome Allison, Michael, and Varshini! CTCF is frequently mutated in cancer and in neurodevelopmental disorders: Jane Skok's lab used an inducible genetic complementation system to study the effects on these mutations on transcription and genome organization, and Christos and Domenic contributed excellent FRAP analyses to understand how these mutations affect CTCF's dynamics, residence time, and bound fraction.
See the full preprint on BioRxiv. |
Archives
July 2024
|