Congratulations to Asmita whose protocol paper is now published in Methods in Molecular Biology.
Asmita wrote this step-by-step protocol to help anyone interested in Single-Particle Tracking (SPT) get started with SPT experiments and how to analyze the data. The focus is on transcription factors in mammalian cells, but we also discuss key considerations to avoid common biases in SPT and how to optimize the experiments in general. Finally, we discuss how to analyze "fastSPT" data with Spot-On. Read the full protocol here: https://link.springer.com/protocol/10.1007%2F978-1-0716-2140-0_9 New preprint: Dynamics of CTCF and cohesin mediated chromatin looping revealed by live-cell imaging2/1/2022
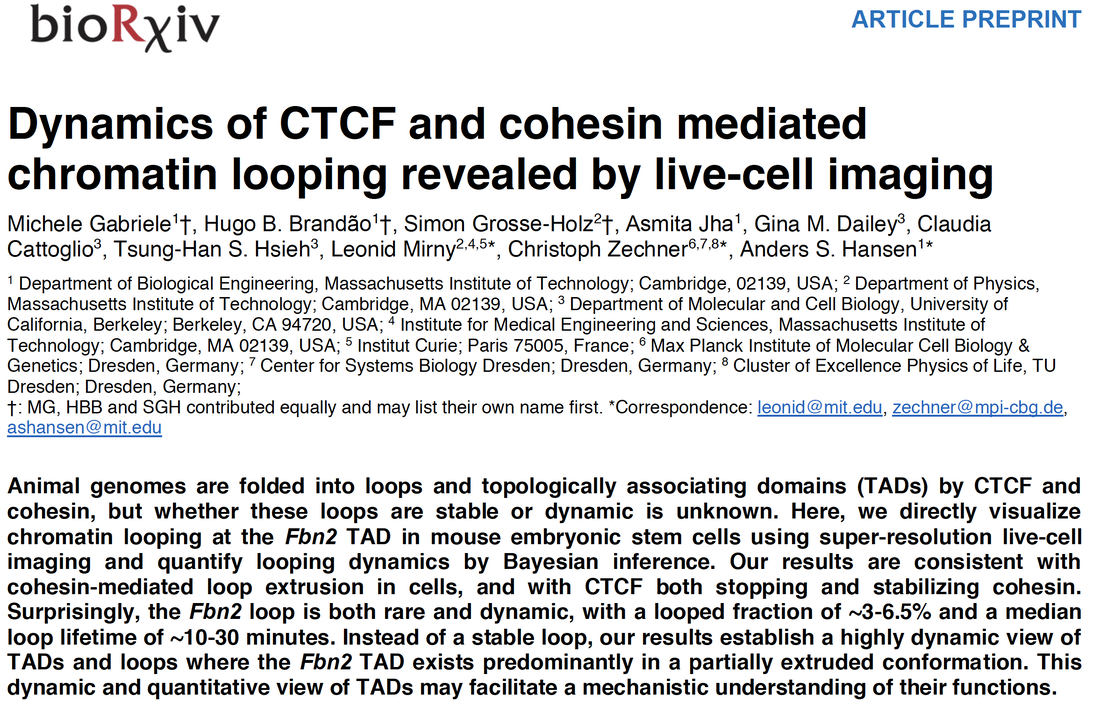
We are thrilled to share our preprint on the dynamics of chromatin looping.
Using 3D Super-Resolution Live-Cell Imaging, we visualize the CTCF/cohesin-mediated loops that hold together TADs and loop domains for the first time in living cells. Surprisingly, these loops are both highly dynamic (median lifetime of ~10-30 min) and very rare (~3-6.5%). Instead of a stable loop, our results establish a highly dynamic view of TADs and loops where TADs exists predominantly in a partially extruded conformation. Read more here. Data here. Code here. This was a team effort and only possible due to the amazing lead from Michele, Hugo, Simon, and Asmita who lead the project and put it all together during a global pandemic! This was a collaborative project with Leonid Mirny (MIT Physics) and Christoph Zechner (Max Planck, Dresden) and the project began in Berkeley in collaboration with Gina Dailey, Claudia Cattoglio, and TH Stanley Hsieh. |
Archives
July 2024
|